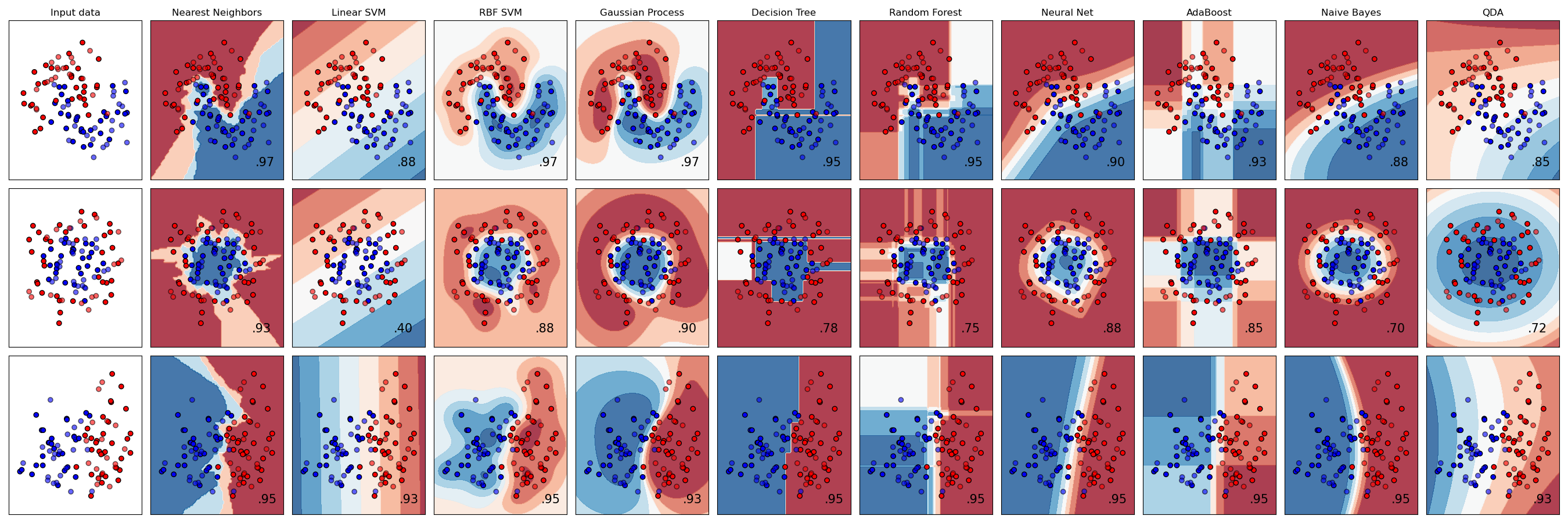
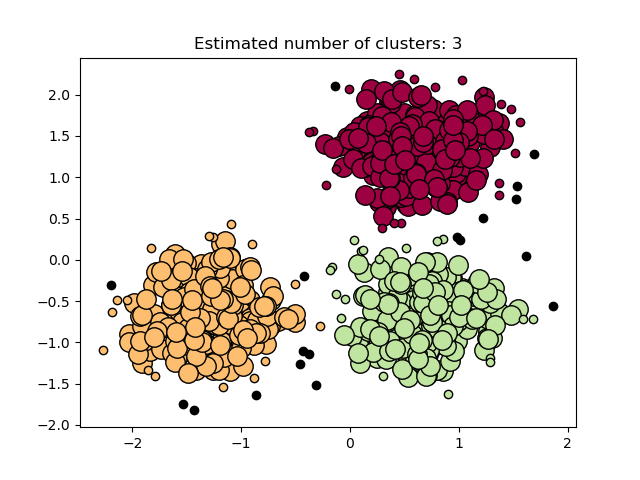
import numpy as np
from sklearn.datasets import fetch_20newsgroups
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
from sklearn.preprocessing import FunctionTransformer
from sklearn.linear_model import SGDClassifier
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.semi_supervised import SelfTrainingClassifier
from sklearn.semi_supervised import LabelSpreading
from sklearn.metrics import f1_score
# Loading dataset containing first five categories
data = fetch_20newsgroups(
subset="train",
categories=[
"alt.atheism",
"comp.graphics",
"comp.os.ms-windows.misc",
"comp.sys.ibm.pc.hardware",
"comp.sys.mac.hardware",
],
)
print("%d documents" % len(data.filenames))
print("%d categories" % len(data.target_names))
print()
# Parameters
sdg_params = dict(alpha=1e-5, penalty="l2", loss="log")
vectorizer_params = dict(ngram_range=(1, 2), min_df=5, max_df=0.8)
# Supervised Pipeline
pipeline = Pipeline(
[
("vect", CountVectorizer(**vectorizer_params)),
("tfidf", TfidfTransformer()),
("clf", SGDClassifier(**sdg_params)),
]
)
# SelfTraining Pipeline
st_pipeline = Pipeline(
[
("vect", CountVectorizer(**vectorizer_params)),
("tfidf", TfidfTransformer()),
("clf", SelfTrainingClassifier(SGDClassifier(**sdg_params), verbose=True)),
]
)
# LabelSpreading Pipeline
ls_pipeline = Pipeline(
[
("vect", CountVectorizer(**vectorizer_params)),
("tfidf", TfidfTransformer()),
# LabelSpreading does not support dense matrices
("todense", FunctionTransformer(lambda x: x.todense())),
("clf", LabelSpreading()),
]
)
def eval_and_print_metrics(clf, X_train, y_train, X_test, y_test):
print("Number of training samples:", len(X_train))
print("Unlabeled samples in training set:", sum(1 for x in y_train if x == -1))
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
print(
"Micro-averaged F1 score on test set: %0.3f"
% f1_score(y_test, y_pred, average="micro")
)
print("-" * 10)
print()
if __name__ == "__main__":
X, y = data.data, data.target
X_train, X_test, y_train, y_test = train_test_split(X, y)
print("Supervised SGDClassifier on 100% of the data:")
eval_and_print_metrics(pipeline, X_train, y_train, X_test, y_test)
# select a mask of 20% of the train dataset
y_mask = np.random.rand(len(y_train)) < 0.2
# X_20 and y_20 are the subset of the train dataset indicated by the mask
X_20, y_20 = map(
list, zip(*((x, y) for x, y, m in zip(X_train, y_train, y_mask) if m))
)
print("Supervised SGDClassifier on 20% of the training data:")
eval_and_print_metrics(pipeline, X_20, y_20, X_test, y_test)
# set the non-masked subset to be unlabeled
y_train[~y_mask] = -1
print("SelfTrainingClassifier on 20% of the training data (rest is unlabeled):")
eval_and_print_metrics(st_pipeline, X_train, y_train, X_test, y_test)
print("LabelSpreading on 20% of the data (rest is unlabeled):")
eval_and_print_metrics(ls_pipeline, X_train, y_train, X_test, y_test)